
Synthetic Biology. We aim to reverse engineer cellular mechanisms regulated by biomolecular components and to use synthetic analogs of those components to program systems with similar capabilities. A hallmark of our work is synthetic cells incorporating adaptive protein/DNA organelles that assemble and disassemble upon environmental cue. Our strategy focuses on thermodynamic design rules that govern liquid-liquid phase separation of intrinsically disordered proteins into architectures that resemble membraneless organelles of living cells.
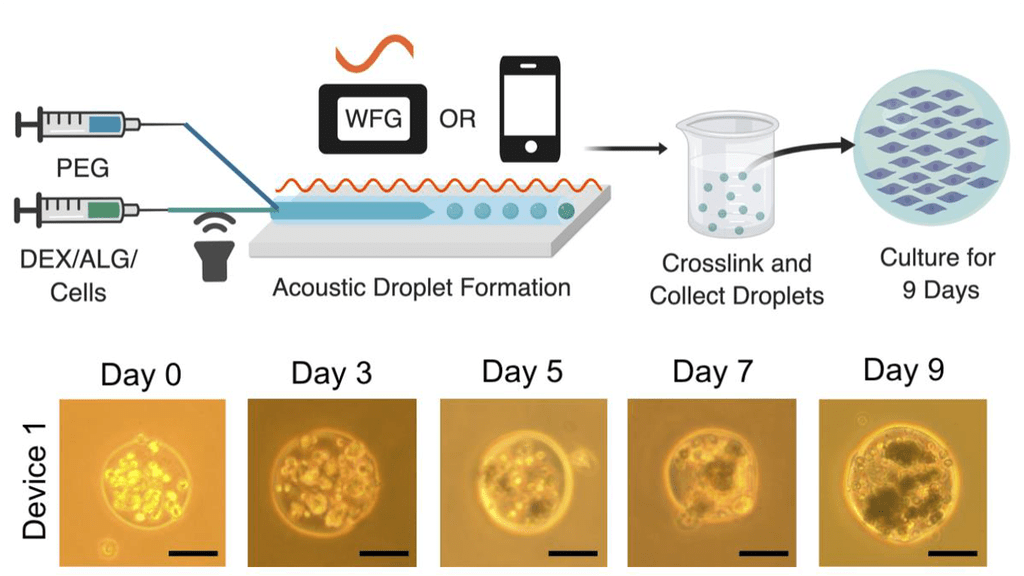
All-Water Bioengineered Systems Segregative liquid phase separation refers to the formation of two immiscible polymeric aqueous liquid phases where two polymers are in a soluble state with water, but phase separate from each other . This leads to an aqueous two-phase system (ATPS), where each phase is enriched in one or the other polymer. By combining ATPS and microfluidic technologies we create water-in-water emulsion droplets for bioengineered platforms. Engineered all-water systems include acoustofluidic fabrication of tumor-like spheroids within hydrogel microspheres and membrane free artificial cell compartments housing nucleoprotein organelles.
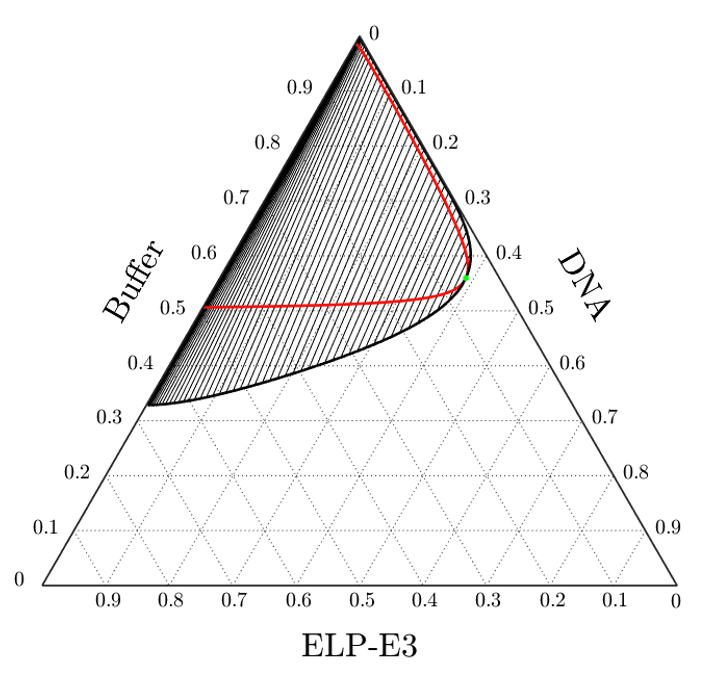
Biophysics. We work to elucidate the thermodynamic design rules that govern liquid-liquid phase separation of intrinsically disordered proteins (IDPs) and nucleic acids into condensed phase spheres known as coacervates. Living cells use protein/RNA/DNA coacervate membrane-free organelles to regulate gene expression in response to environment by phase separating and physically sequestering regulatory biomolecules. Using theoretical analyses, we interpret the phase behavior of archetypal IDP and DNA sequences to assemble synthetic organelles. Measured thermodynamic interaction parameters define the energetics of pairwise polymer-solvent and polymer-polymer interactions that establish multicomponent free energy landscapes to demarcate protein and nucleic acid amounts in each synthetic compartment.
